Download Stable MCCE executable for LINUX or Mac OSX:
Photosystem II
Photosystem II (PSII) is the photosynthetic protein that takes electrons from water and releases protons into the lumen (on the P-side of the chloroplasts). The reaction releases O2 and is the source of almost all the O2 in the atmosphere Water oxidation takes place at the catalytic site in PSII known as Oxygen Evolving Complex (OEC). The movement of protons and water takes place in water channels near the OEC. My work combines computational methodologies Molecular Dynamics (MD) and MCCE (Multi Conformation Continuum Electrostatics) to understand the mechanism of protons and water dynamics in wild-type PSII and mutants. Classical MD provides the conformational sampling of protein with time. MCCE is also a classical based method that provides an insight about the protonation state changes of the residues upon the water-splitting process.

Complex I
Complex I, NADH-ubiquinone oxidoreductase, the first enzyme in the respiratory chain, contributes almost 40% of the energy stored in the respiratory chain of bacteria and mitochondria. It works by pumping four protons through transiently open pathways from N- to P-side driven by the transfer of electrons from NADH to a quinone. To understands to proton pathway in complex I, we are applying the grand canonical Monte Carlo simulations (MCCE) to snapshots from large-scale molecular dynamics trajectories. Our hydrogen bond analysis technique helps us find the proton pumping pathway. MCCE is also being used to find the protonation states of titrable residues in different intermediates in the reaction cycle to see where they are transiently held as they cross the membrane.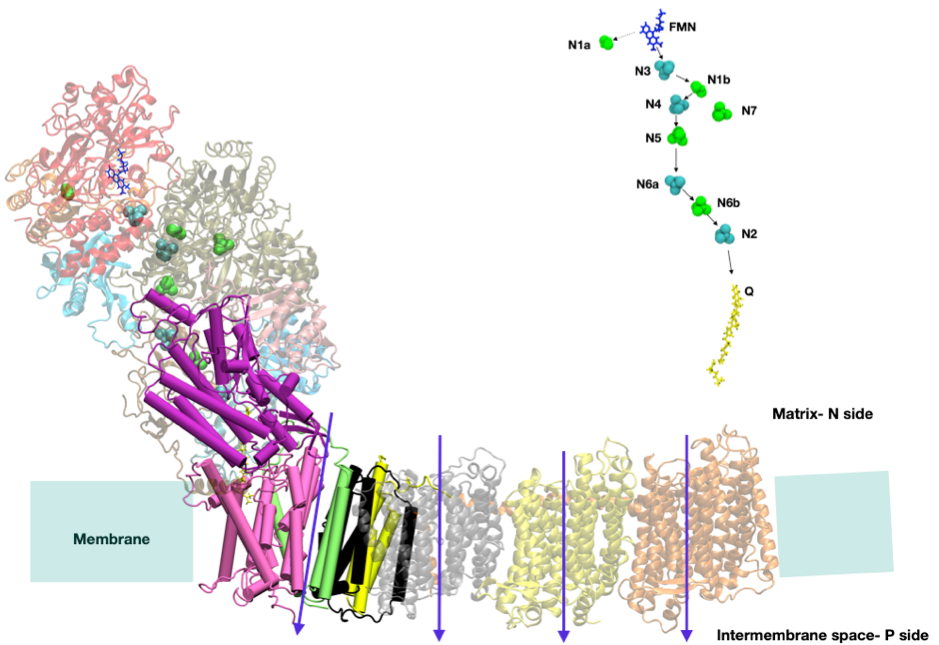
Cytochrome c oxidase
CcO (complex IV) is the terminal protein of the respiratory electron transfer chain in mitochondria and aerobic bacteria. It uses the electrons from cytochrome c produced in complex III, to reduce O2 to water. CcO catalyzes the reaction at the active site Binuclear Complex (BNC), which contains a heme (heme a3) and a copper (CuB). During one catalytic cycle, four electrons are transferred from cytochromes c bound at the P side to CuA, a bi-copper site, and then to a low-spin heme a, finally to the BNC. Meanwhile, eight protons are taken up from N-side, with four protons delivered to the BNC as needed for the oxygen chemistry (chemical protons), and four protons translocated across the membrane (pumped protons). We are studying the proton pumping mechanism of Cytochrome c Oxidase through MCCE and its hydrogen bond network module. Residues that are highly hydrogen-bonded and the inter-cluster connection reveal the proton pumping pathway on Cytochrome c Oxidase.
Gramicidin
We are looking at modeling explicit waters inside the Gramicidin A channel with Monte Carlo sampling. We are comparing the water orientation in MD trajectories (with classical force fields and with a polarized force field_ and with MC simulation (using MCCE). The water orientation in the standard MCCE calculations behaves more like the water in the polarized force field. We are using MCCE to estimate the barrier for moving hydronium through the gA channel. 
